Note
Go to the end to download the full example code.
U-NO on Darcy-Flow
In this example, we demonstrate how to train a U-shaped Neural Operator on the small Darcy-Flow example we ship with the package
import torch
import matplotlib.pyplot as plt
import sys
from neuralop.models import UNO
from neuralop import Trainer
from neuralop.training import AdamW
from neuralop.data.datasets import load_darcy_flow_small
from neuralop.utils import count_model_params
from neuralop import LpLoss, H1Loss
device = 'cpu'
Loading the Darcy Flow dataset
train_loader, test_loaders, data_processor = load_darcy_flow_small(
n_train=1000, batch_size=32,
test_resolutions=[16, 32], n_tests=[100, 50],
test_batch_sizes=[32, 32],
)
model = UNO(in_channels=1,
out_channels=1,
hidden_channels=64,
projection_channels=64,
uno_out_channels=[32,64,64,64,32],
uno_n_modes=[[16,16],[8,8],[8,8],[8,8],[16,16]],
uno_scalings=[[1.0,1.0],[0.5,0.5],[1,1],[2,2],[1,1]],
horizontal_skips_map=None,
channel_mlp_skip="linear",
n_layers = 5,
domain_padding=0.2)
model = model.to(device)
n_params = count_model_params(model)
print(f'\nOur model has {n_params} parameters.')
sys.stdout.flush()
/home/runner/work/neuraloperator/neuraloperator/neuralop/data/datasets/pt_dataset.py:93: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
data = torch.load(
Loading test db for resolution 16 with 100 samples
/home/runner/work/neuraloperator/neuraloperator/neuralop/data/datasets/pt_dataset.py:172: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
data = torch.load(Path(root_dir).joinpath(f"{dataset_name}_test_{res}.pt").as_posix())
Loading test db for resolution 32 with 50 samples
fno_skip='linear'
channel_mlp_skip='linear'
fno_skip='linear'
channel_mlp_skip='linear'
fno_skip='linear'
channel_mlp_skip='linear'
fno_skip='linear'
channel_mlp_skip='linear'
fno_skip='linear'
channel_mlp_skip='linear'
Our model has 2700097 parameters.
Create the optimizer
optimizer = AdamW(model.parameters(),
lr=8e-3,
weight_decay=1e-4)
scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=30)
Creating the losses
l2loss = LpLoss(d=2, p=2)
h1loss = H1Loss(d=2)
train_loss = h1loss
eval_losses={'h1': h1loss, 'l2': l2loss}
print('\n### MODEL ###\n', model)
print('\n### OPTIMIZER ###\n', optimizer)
print('\n### SCHEDULER ###\n', scheduler)
print('\n### LOSSES ###')
print(f'\n * Train: {train_loss}')
print(f'\n * Test: {eval_losses}')
sys.stdout.flush()
### MODEL ###
UNO(
(positional_embedding): GridEmbeddingND()
(domain_padding): DomainPadding()
(lifting): ChannelMLP(
(fcs): ModuleList(
(0): Conv1d(3, 256, kernel_size=(1,), stride=(1,))
(1): Conv1d(256, 64, kernel_size=(1,), stride=(1,))
)
)
(fno_blocks): ModuleList(
(0): FNOBlocks(
(convs): ModuleList(
(0): SpectralConv(
(weight): DenseTensor(shape=torch.Size([64, 32, 16, 9]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(64, 32, kernel_size=(1,), stride=(1,), bias=False)
)
)
(channel_mlp): ModuleList(
(0): ChannelMLP(
(fcs): ModuleList(
(0): Conv1d(32, 16, kernel_size=(1,), stride=(1,))
(1): Conv1d(16, 32, kernel_size=(1,), stride=(1,))
)
)
)
(channel_mlp_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(64, 32, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(1): FNOBlocks(
(convs): ModuleList(
(0): SpectralConv(
(weight): DenseTensor(shape=torch.Size([32, 64, 8, 5]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(32, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
(channel_mlp): ModuleList(
(0): ChannelMLP(
(fcs): ModuleList(
(0): Conv1d(64, 32, kernel_size=(1,), stride=(1,))
(1): Conv1d(32, 64, kernel_size=(1,), stride=(1,))
)
)
)
(channel_mlp_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(32, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(2): FNOBlocks(
(convs): ModuleList(
(0): SpectralConv(
(weight): DenseTensor(shape=torch.Size([64, 64, 8, 5]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(64, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
(channel_mlp): ModuleList(
(0): ChannelMLP(
(fcs): ModuleList(
(0): Conv1d(64, 32, kernel_size=(1,), stride=(1,))
(1): Conv1d(32, 64, kernel_size=(1,), stride=(1,))
)
)
)
(channel_mlp_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(64, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(3): FNOBlocks(
(convs): ModuleList(
(0): SpectralConv(
(weight): DenseTensor(shape=torch.Size([128, 64, 8, 5]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(128, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
(channel_mlp): ModuleList(
(0): ChannelMLP(
(fcs): ModuleList(
(0): Conv1d(64, 32, kernel_size=(1,), stride=(1,))
(1): Conv1d(32, 64, kernel_size=(1,), stride=(1,))
)
)
)
(channel_mlp_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(128, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(4): FNOBlocks(
(convs): ModuleList(
(0): SpectralConv(
(weight): DenseTensor(shape=torch.Size([96, 32, 16, 9]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(96, 32, kernel_size=(1,), stride=(1,), bias=False)
)
)
(channel_mlp): ModuleList(
(0): ChannelMLP(
(fcs): ModuleList(
(0): Conv1d(32, 16, kernel_size=(1,), stride=(1,))
(1): Conv1d(16, 32, kernel_size=(1,), stride=(1,))
)
)
)
(channel_mlp_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(96, 32, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
)
(horizontal_skips): ModuleDict(
(0): Flattened1dConv(
(conv): Conv1d(32, 32, kernel_size=(1,), stride=(1,), bias=False)
)
(1): Flattened1dConv(
(conv): Conv1d(64, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
(projection): ChannelMLP(
(fcs): ModuleList(
(0): Conv1d(32, 64, kernel_size=(1,), stride=(1,))
(1): Conv1d(64, 1, kernel_size=(1,), stride=(1,))
)
)
)
### OPTIMIZER ###
AdamW (
Parameter Group 0
betas: (0.9, 0.999)
correct_bias: True
eps: 1e-06
initial_lr: 0.008
lr: 0.008
weight_decay: 0.0001
)
### SCHEDULER ###
<torch.optim.lr_scheduler.CosineAnnealingLR object at 0x7fb4d44d4910>
### LOSSES ###
* Train: <neuralop.losses.data_losses.H1Loss object at 0x7fb4d69a5d30>
* Test: {'h1': <neuralop.losses.data_losses.H1Loss object at 0x7fb4d69a5d30>, 'l2': <neuralop.losses.data_losses.LpLoss object at 0x7fb4d44d6990>}
Create the trainer
trainer = Trainer(model=model,
n_epochs=20,
device=device,
data_processor=data_processor,
wandb_log=False,
eval_interval=3,
use_distributed=False,
verbose=True)
Actually train the model on our small Darcy-Flow dataset
trainer.train(train_loader=train_loader,
test_loaders=test_loaders,
optimizer=optimizer,
scheduler=scheduler,
regularizer=False,
training_loss=train_loss,
eval_losses=eval_losses)
Training on 1000 samples
Testing on [50, 50] samples on resolutions [16, 32].
Raw outputs of shape torch.Size([32, 1, 16, 16])
[0] time=10.18, avg_loss=0.6187, train_err=19.3356
Eval: 16_h1=0.6084, 16_l2=0.4112, 32_h1=0.8841, 32_l2=0.6583
[3] time=10.10, avg_loss=0.2388, train_err=7.4617
Eval: 16_h1=0.2834, 16_l2=0.2026, 32_h1=0.8187, 32_l2=0.5866
[6] time=10.15, avg_loss=0.2374, train_err=7.4177
Eval: 16_h1=0.2556, 16_l2=0.1647, 32_h1=0.8075, 32_l2=0.5393
[9] time=10.09, avg_loss=0.2171, train_err=6.7847
Eval: 16_h1=0.2840, 16_l2=0.1783, 32_h1=0.8275, 32_l2=0.5473
[12] time=10.19, avg_loss=0.2142, train_err=6.6923
Eval: 16_h1=0.2434, 16_l2=0.1549, 32_h1=0.7687, 32_l2=0.4845
[15] time=10.09, avg_loss=0.1608, train_err=5.0265
Eval: 16_h1=0.2652, 16_l2=0.1628, 32_h1=0.8072, 32_l2=0.4911
[18] time=10.11, avg_loss=0.1565, train_err=4.8909
Eval: 16_h1=0.2503, 16_l2=0.1604, 32_h1=0.7746, 32_l2=0.5099
{'train_err': 3.99467795714736, 'avg_loss': 0.12782969462871552, 'avg_lasso_loss': None, 'epoch_train_time': 10.095863415000053}
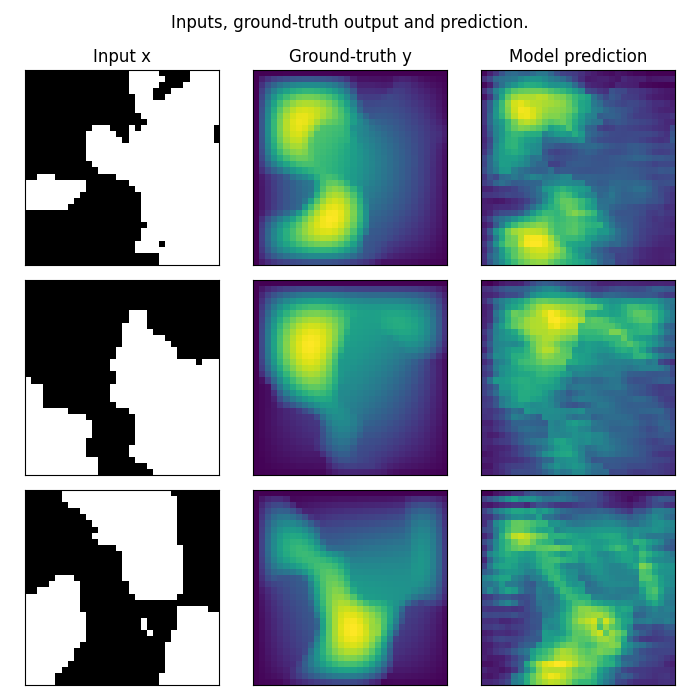
Plot the prediction, and compare with the ground-truth Note that we trained on a very small resolution for a very small number of epochs In practice, we would train at larger resolution, on many more samples.
However, for practicity, we created a minimal example that i) fits in just a few Mb of memory ii) can be trained quickly on CPU
In practice we would train a Neural Operator on one or multiple GPUs
test_samples = test_loaders[32].dataset
fig = plt.figure(figsize=(7, 7))
for index in range(3):
data = test_samples[index]
data = data_processor.preprocess(data, batched=False)
# Input x
x = data['x']
# Ground-truth
y = data['y']
# Model prediction
out = model(x.unsqueeze(0).to(device)).cpu()
ax = fig.add_subplot(3, 3, index*3 + 1)
ax.imshow(x[0], cmap='gray')
if index == 0:
ax.set_title('Input x')
plt.xticks([], [])
plt.yticks([], [])
ax = fig.add_subplot(3, 3, index*3 + 2)
ax.imshow(y.squeeze())
if index == 0:
ax.set_title('Ground-truth y')
plt.xticks([], [])
plt.yticks([], [])
ax = fig.add_subplot(3, 3, index*3 + 3)
ax.imshow(out.squeeze().detach().numpy())
if index == 0:
ax.set_title('Model prediction')
plt.xticks([], [])
plt.yticks([], [])
fig.suptitle('Inputs, ground-truth output and prediction.', y=0.98)
plt.tight_layout()
fig.show()
Total running time of the script: (3 minutes 25.541 seconds)