Note
Go to the end to download the full example code
U-NO on Darcy-Flow
In this example, we demonstrate how to train a U-shaped Neural Operator on the small Darcy-Flow example we ship with the package
import torch
import matplotlib.pyplot as plt
import sys
from neuralop.models import TFNO, UNO
from neuralop import Trainer
from neuralop.datasets import load_darcy_flow_small
from neuralop.utils import count_model_params
from neuralop import LpLoss, H1Loss
device = 'cpu'
Loading the Darcy Flow dataset
train_loader, test_loaders, data_processor = load_darcy_flow_small(
n_train=1000, batch_size=32,
test_resolutions=[16, 32], n_tests=[100, 50],
test_batch_sizes=[32, 32],
)
model = UNO(3,1, hidden_channels=64, projection_channels=64,uno_out_channels = [32,64,64,64,32], \
uno_n_modes= [[16,16],[8,8],[8,8],[8,8],[16,16]], uno_scalings= [[1.0,1.0],[0.5,0.5],[1,1],[2,2],[1,1]],\
horizontal_skips_map = None, n_layers = 5, domain_padding = 0.2)
model = model.to(device)
n_params = count_model_params(model)
print(f'\nOur model has {n_params} parameters.')
sys.stdout.flush()
Loading test db at resolution 32 with 50 samples and batch-size=32
Our model has 2665921 parameters.
Create the optimizer
optimizer = torch.optim.Adam(model.parameters(),
lr=8e-3,
weight_decay=1e-4)
scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=30)
Creating the losses
l2loss = LpLoss(d=2, p=2)
h1loss = H1Loss(d=2)
train_loss = h1loss
eval_losses={'h1': h1loss, 'l2': l2loss}
print('\n### MODEL ###\n', model)
print('\n### OPTIMIZER ###\n', optimizer)
print('\n### SCHEDULER ###\n', scheduler)
print('\n### LOSSES ###')
print(f'\n * Train: {train_loss}')
print(f'\n * Test: {eval_losses}')
sys.stdout.flush()
### MODEL ###
UNO(
(domain_padding): DomainPadding()
(lifting): MLP(
(fcs): ModuleList(
(0): Conv1d(3, 256, kernel_size=(1,), stride=(1,))
(1): Conv1d(256, 64, kernel_size=(1,), stride=(1,))
)
)
(fno_blocks): ModuleList(
(0): FNOBlocks(
(convs): SpectralConv(
(weight): ModuleList(
(0): ComplexDenseTensor(shape=torch.Size([64, 32, 16, 9]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(64, 32, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(1): FNOBlocks(
(convs): SpectralConv(
(weight): ModuleList(
(0): ComplexDenseTensor(shape=torch.Size([32, 64, 8, 5]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(32, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(2): FNOBlocks(
(convs): SpectralConv(
(weight): ModuleList(
(0): ComplexDenseTensor(shape=torch.Size([64, 64, 8, 5]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(64, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(3): FNOBlocks(
(convs): SpectralConv(
(weight): ModuleList(
(0): ComplexDenseTensor(shape=torch.Size([128, 64, 8, 5]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(128, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
(4): FNOBlocks(
(convs): SpectralConv(
(weight): ModuleList(
(0): ComplexDenseTensor(shape=torch.Size([96, 32, 16, 9]), rank=None)
)
)
(fno_skips): ModuleList(
(0): Flattened1dConv(
(conv): Conv1d(96, 32, kernel_size=(1,), stride=(1,), bias=False)
)
)
)
)
(horizontal_skips): ModuleDict(
(0): Flattened1dConv(
(conv): Conv1d(32, 32, kernel_size=(1,), stride=(1,), bias=False)
)
(1): Flattened1dConv(
(conv): Conv1d(64, 64, kernel_size=(1,), stride=(1,), bias=False)
)
)
(projection): MLP(
(fcs): ModuleList(
(0): Conv1d(32, 64, kernel_size=(1,), stride=(1,))
(1): Conv1d(64, 1, kernel_size=(1,), stride=(1,))
)
)
)
### OPTIMIZER ###
Adam (
Parameter Group 0
amsgrad: False
betas: (0.9, 0.999)
capturable: False
differentiable: False
eps: 1e-08
foreach: None
fused: None
initial_lr: 0.008
lr: 0.008
maximize: False
weight_decay: 0.0001
)
### SCHEDULER ###
<torch.optim.lr_scheduler.CosineAnnealingLR object at 0x7f1cd6270c40>
### LOSSES ###
* Train: <neuralop.losses.data_losses.H1Loss object at 0x7f1cd62700d0>
* Test: {'h1': <neuralop.losses.data_losses.H1Loss object at 0x7f1cd62700d0>, 'l2': <neuralop.losses.data_losses.LpLoss object at 0x7f1cd6270be0>}
Create the trainer
trainer = Trainer(model=model,
n_epochs=20,
device=device,
data_processor=data_processor,
wandb_log=False,
log_test_interval=3,
use_distributed=False,
verbose=True)
self.override_load_to_device=False
self.overrides_loss=False
Actually train the model on our small Darcy-Flow dataset
trainer.train(train_loader=train_loader,
test_loaders=test_loaders,
optimizer=optimizer,
scheduler=scheduler,
regularizer=False,
training_loss=train_loss,
eval_losses=eval_losses)
{'32_h1': 0.8166693496704102, '32_l2': 0.839678955078125}
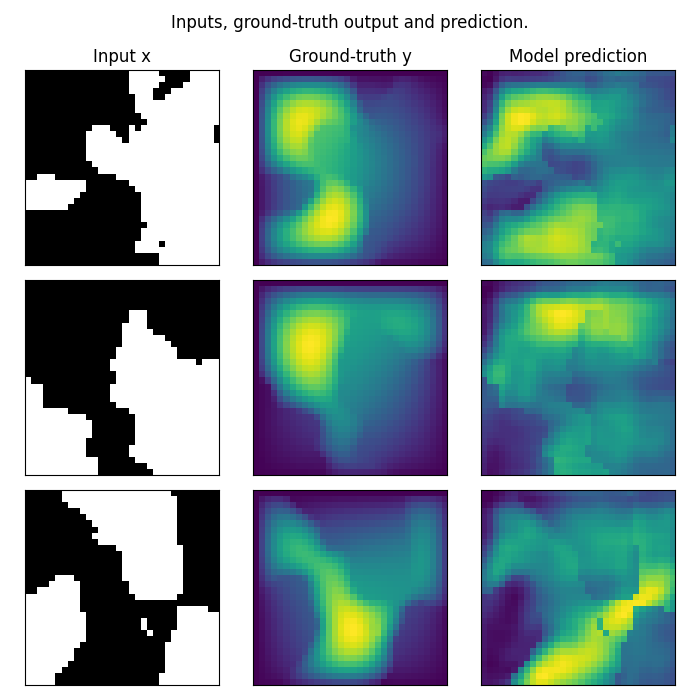
Plot the prediction, and compare with the ground-truth Note that we trained on a very small resolution for a very small number of epochs In practice, we would train at larger resolution, on many more samples.
However, for practicity, we created a minimal example that i) fits in just a few Mb of memory ii) can be trained quickly on CPU
In practice we would train a Neural Operator on one or multiple GPUs
test_samples = test_loaders[32].dataset
fig = plt.figure(figsize=(7, 7))
for index in range(3):
data = test_samples[index]
data = data_processor.preprocess(data, batched=False)
# Input x
x = data['x']
# Ground-truth
y = data['y']
# Model prediction
out = model(x.unsqueeze(0).to(device)).cpu()
ax = fig.add_subplot(3, 3, index*3 + 1)
ax.imshow(x[0], cmap='gray')
if index == 0:
ax.set_title('Input x')
plt.xticks([], [])
plt.yticks([], [])
ax = fig.add_subplot(3, 3, index*3 + 2)
ax.imshow(y.squeeze())
if index == 0:
ax.set_title('Ground-truth y')
plt.xticks([], [])
plt.yticks([], [])
ax = fig.add_subplot(3, 3, index*3 + 3)
ax.imshow(out.squeeze().detach().numpy())
if index == 0:
ax.set_title('Model prediction')
plt.xticks([], [])
plt.yticks([], [])
fig.suptitle('Inputs, ground-truth output and prediction.', y=0.98)
plt.tight_layout()
fig.show()
Total running time of the script: (1 minutes 56.484 seconds)